Risk Prediction Method for Immunoglobulin-E-Level-Dependent Diseases in Human Blood Serum
In the present-day world, the study of genetic information becomes more and more important, which generated a novel approach to diagnostics and treatment of different diseases in personalized medicine. The advancements in technology such as next-generation sequencing (NGS) and DNA microarrays enabled reducing genome information deciphering time. Nevertheless, their application to routine lab diagnostics seems to be unlikely because of inevitable great investments in equipment and consumables, as well as a considerable amount of time, high risk of sample contamination and deep expertise of researchers. These limitations can be avoided when using the TaqMan method. Its characteristics allow elaborating accurate and low-cost systems for individual risk prediction of diseases related to different single-nucleotide polymorphisms (SNPs). The systems of this kind do not require either specialized knowledge to perform reactions or expensive equipment and consumables, and their results do not need any additional processing.
Nowadays, one of the most burning issues is allergic diseases. Statistics indicate that they are placed first in the disease incidence for all age groups. Considering this, it is important to focus on the predisposition diagnostics, even at the preclinical stage, which will make it possible to initiate early prevention and health care control in high-risk groups. At the moment, the past medical history from the patient’s family members is the main way of allergy risk estimation. The disadvantage of this method is its subjectivity. Our recently developed complex analysis system of genetic polymorphisms related to allergic diseases will increase the number of available methods and improve the risk estimation precision.
Investigating evolutions of allergic reactions related to Type I hypersensitivity, the immunoglobulin E (IgE) was discovered. This type of hypersensitivity, also called anaphylactic, causes, after contacting a primary allergen, a reaction cascade due to the specific IgE synthesis by plasmacytes, which finally results in allergic clinical manifestations. The IgE played the crucial role in the evolution protecting the organism against parasites, extremely relevant for human survival in the wild. The parasite infection spread, however, decreased with the progress of civilization, but the level of the serum IgE is still high, and this is why contacts with allergens started to cause pathological reactions. Later, some studies pointed out the correlation between the serum IgE level and several diseases. At high serum IgE levels, there is an increasing risk of such diseases as atopic dermatitis, chronical urticaria, asthma, allergic rhinitis, allergic rhinoconjunctivitis. And vice versa, a tendency to parasitic diseases and animal toxins is believed to relate to the lower serum IgE level. Knowing this, we can assert that the high serum IgE level is a marker for several allergic disorders, and the low serum IgE level is one of the causes for weakened immunity to parasitic diseases and animal toxins.
We faced the challenge to elaborate a system of TaqMan probes and primers to analyze human polymorphisms rs2251746 and rs2427837 using allelic discrimination. The rs2251746 and rs2427837 single nucleotide polymorphisms occur in the gene coding for the alpha chain of the high-affinity IgE receptor (FCER1A) on the 1q23 chromosome. Various studies demonstrated a strong correlation between the rs2251746 and rs2427837 presence in the human genome and the serum IgE level, and also allergic reaction risks. This system enables estimating the risks of human diseases dependent on the serum IgE level, such as atopic dermatitis, chronical urticaria, atopic eczema, asthma, allergic rhinitis, allergic conjunctivitis and allergic rhinoconjunctivitis.
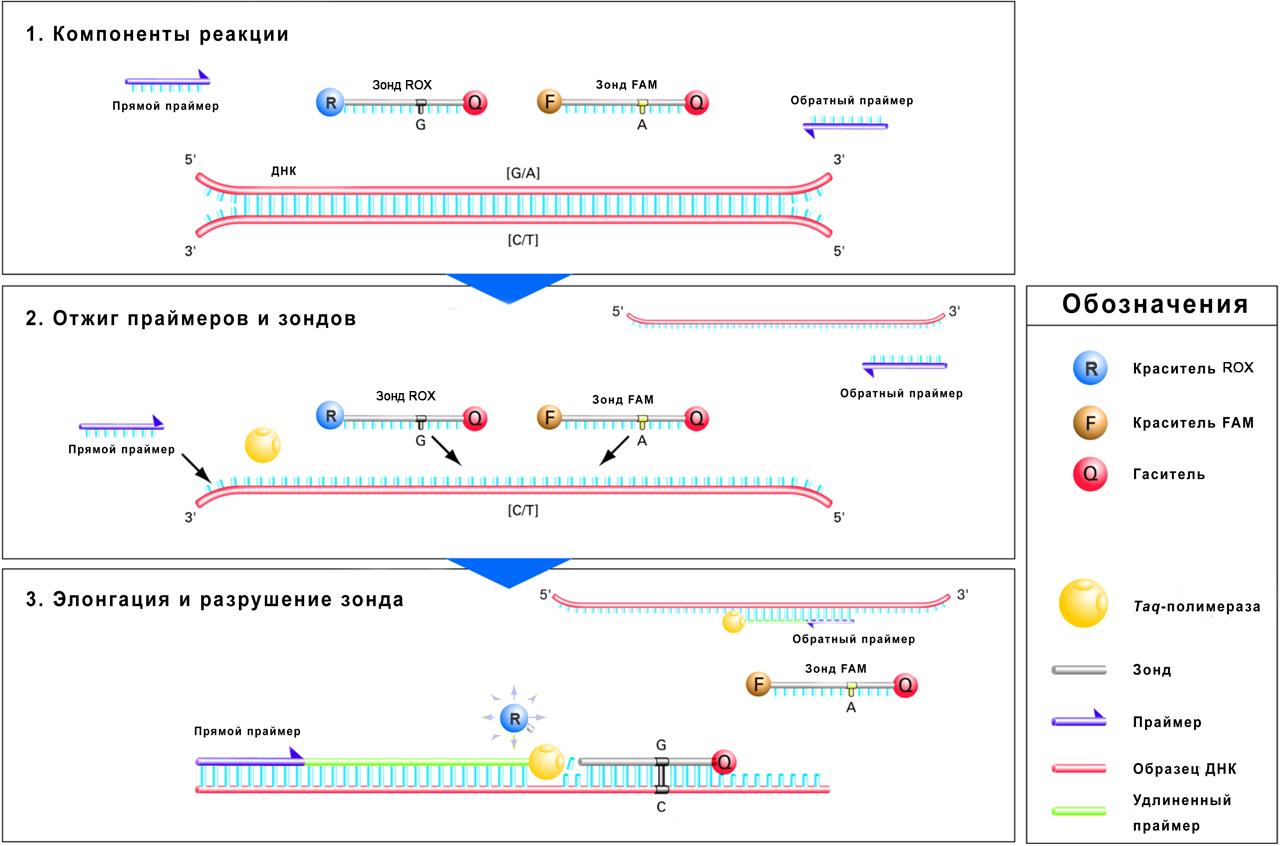
The TaqMan assay is widely used in allelic discrimination due to its sensitivity and specificity.
The TaqMan probe is an allele-specific oligonucleotide probe labelled by a fluorescent dye on the 5’ end and by a quencher on the 3’ end.
The method relies on the 5’-3’ exonuclease activity of the Taq DNA polymerase able to cleave the 5’-end nucleotides of the double-stranded DNA releasing mono- and oligonucleotides.
If a probe is not fully complementary to DNA, it is not cleaved by the Taq polymerase, but entirely detached from the nucleotide sequence, and this is why the fluorescence from the fluorescent dye on the 5’ end will be completely absorbed by the quencher on the 3’ end of the probe due to the fluorescence resonance energy transfer (FRET).
If a probe is fully complementary to DNA, it is cleaved by the Taq polymerase, the probe breaks into pieces, and the fluorescent dye and the quencher are released into the solution. Since they are not bound anymore, the quencher effect disappears, and the fluorescence from the fluorophore can be identified by the real-time amplifier. The fluorescence intensity will be proportional to the amount of polymerase chain reaction (PCR) products.
Allelic discrimination is a multiplex (more than one primer/probe pairs for a reaction) end-point (data readout is performed at the end of the PCR process) method implied for differentiation between genotypes, mutations and polymorphisms by comparing the fluorescence obtained using dye labelled probes. Applying two pairs of primers and probes in every reaction mixture enables genotyping two possible variants of one SNP in the targeted sequence. However, the actual amount of the PCR product is not detected, which makes allelic discrimination a reliable assay.
Special dyes employed for each allele variant provide that the luminescence of these dyes differs.
Genotyping was conducted in relative fluorescence units (RFUs). For rs2251746, the probe with a FAM fluorescent dye corresponds to the T allele, the ROX dye probe to the C allele. For rs2427837, the probe with a FAM fluorescent dye corresponds to the A allele, the ROX dye probe to the G allele. Genotype identification for each sample is rather effortless since three distinct clusters of the samples can be identified on the graphs: homozygotes GG/CC, heterozygotes AG/CT, homozygotes AA/TT.
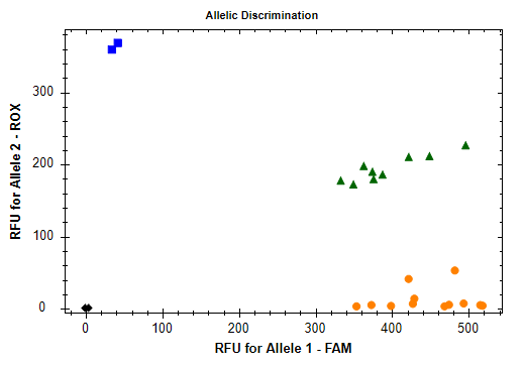 Figure 2. Allelic discrimination of the rs2251746 polymorphism by the TaqMan probe detection method using the RFUs of each probe on the CFX96 amplifier with a real-time detection system: ♦ - negative control, █ - CC homozygotes, ▲ - CT heterozygotes, ● - TT homozygotes.
Figure 2. Allelic discrimination of the rs2251746 polymorphism by the TaqMan probe detection method using the RFUs of each probe on the CFX96 amplifier with a real-time detection system: ♦ - negative control, █ - CC homozygotes, ▲ - CT heterozygotes, ● - TT homozygotes.
Figure 3. Allelic discrimination of the rs2427837 polymorphism by the TaqMan probe detection method using the RFUs of each probe on the CFX96 amplifier with a real-time detection system: ♦ - negative control, █ - GG homozygotes, ▲ - AG heterozygotes, ● - AA homozygotes.
For interpreting the results, you can use the following table:
|
rs2427837 rs2251746 |
GG |
AG |
AA |
|
TT |
high |
higher than normal |
normal |
|
CT |
higher than normal |
normal |
lower than normal |
|
CC |
normal |
lower than normal |
low |
All the results were verified by sequencing.
This method has the following advantages:
1. It allows encompassing a wide range of IgE-dependent diseases.
2. It is accurate, easy-to-use, fast and inexpensive.
3. It does not require any medical knowledge, the past medical patient history and presence of a patient.
4. It can be performed using different types of biological material containing DNA of a patient (venous blood, buccal epithelium, hair follicle etc.).
5. It can be used for any population group.
6. It permits estimating the probability of transmission of this trait to descendants.
7. The assay result does not depend on lifestyle and health status.
Following this research direction, the TaqMan system of probes and primers which identifies an inherited variant of one of the important genes determining the tendency to human longevity, the FOXO3a gene, was designed in the Section of Molecular Genetics of the Cell at DLNP.
Anastasia Ivanova, engineer, Section of Molecular Genetics of the Cell, DLNP






